Funding through Cancer Grand Challenge
We are super excited to be part of team KOODAC, which is funded by the Cancer Grant Challenge initiative to identify and develop small-molecule degraders against oncoproteins that drive solid tumors in children.
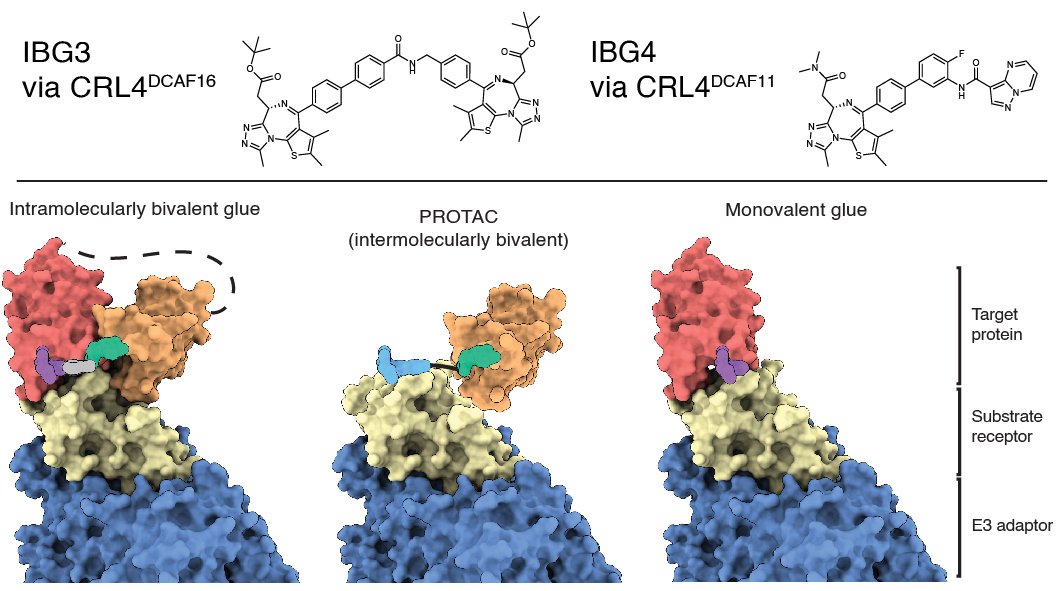
Intramolecular Bivalent Glue Degrader paper out in Nature.
We are very happy that our collaborative work with the Ciulli lab in Dundee has been published in Nature. Congratulations to co-first author Matthias Hinterndorfer.
Intramolecular bivalent glues are a new and differentiated modality to induce targeted protein degradation
New Manuscript in Nature Communications!
Our work shows that small molecules that were previously implied in the induction of autophagy are, when incorporated into heterobifunctional ligands, acting as PROTACs that co-opt the E3 ubiquitin ligase DCAF11.
New Manuscript!
Congratulations to Amanda, Fabian and Jose for their manuscript on glue degrader discovery via morphologic profiling!
Welcome, David!
David Hoi joins the lab as our latest postdoc. Find out more int he Members section.
Meet the Flintstones
We had fun at the legendary, annual CeMM Halloween Party! (and won the costume competition, as expected)
Welcome, Lisa
We are excited to welcome Lisa Kainacher as a new PhD student in the lab. Find more about Lisa in the "lab members” section.
New Grants
We are very grateful for funding of two novel grant applications from the Austrian Science Fund, allowing us to study hydrophobic tagging as well as how proteolytic networks downstream of KRAS can be functionally hijacked with small molecules.
EFMC Award
Georg got the EFMC Prize for Young Medicinal Chemist or Chemical Biologist in Academia for his achievements in targeted protein degradation with molecular glue degraders
New preprint - we identify & mechanistically characterise “Orpinolide”
In another collaboration with the Waldmann lab, we uncover & mechanistically explain the anti-leukemic activity of “Orpinolide”, a novel natural product-like molecule.
Tetrahedron Young Investigator Award
Georg got this prestigious international award for his contributions to the field of targeted protein degradation
A deeper look at ATTECs - a novel preprint out
A 2019 Nature paper reported “ATTECs”: compounds tethering LC3 to mutant huntington to induce its clearance via autophagy. Follow-up studies suggested that this chemotype can yield “AUTACs” that can be engineered to induce the targeted clearance of other proteins targets.
With the Waldmann lab from the MPI in Dortmund, we investigated these compounds with time controlled, FACS-based CRISPR screens, and made the surprising observation that degradation of these heterobifunctional ligands in fact depends on the CRL ligase DCAF11.
Great job by Matthias, Marko and Hana.
Bivalent, intramolecular gluing - a new avenue to targeted protein degradation and proximity-inducing drugs?
In our recent paper with the laboratory of Alessio Ciulli in Dundee, we unravel a novel modality of targeted protein degradation: Intramolecular gluing of protein domains to generate a target-E3 ligase interaction surface. The manuscript is available at biorxiv (link below) and includes functional genomics, biophysical and structural characterization.
Could this be a differentiated strategy to pursue historically challenging multi-domain targets?
Perspective on future of targeted protein degradation out in JACS
Miquel Duran-Frigola, Marko and Georg wrote up their thoughts how AI/ML and multi-omics technologies can help push the boundaries of Targeted Protein Degradation
Ligase tracing manuscript published in JACS
We are happy that our manuscript on E3-ligase specific glue discovery appeared in JACS. Thanks to funding from the FWF, this study is published open-access. Congratulations to first author Alex Hanzl and the entire team.
New manuscript published in Nature Chemical Biology
Our manuscript on mapping functional E3 ligase hotspots is now published. Congratulations to PhD student Alex Hanzl for his first first-author paper!
New preprint on biorxiv!
We recently uploaded a manuscripts that details a novel discovery approach to find molecular glue degraders in a ligase-specific, yet target -agnostic manner.
New team members!
We are delighted to welcome Caroline , the latest PhD students to join the team. Learn more about her background in the Lab Members section!
New preprint on biorxiv
We are excited to share our latest study where we combine functional genomics with biophysics and structural biology to map functional hotspots in E3 ubiquitin ligases that are hijacked by PROTACs and molecular glues. Interestingly, these hotspots might be of clinical relevance since we find that they are disrupted by mutations in patients relapsing from IMiD treatment
Recently, Georg gave a talk at the Targets 2035 webinar series, which is now available on youtube.
New Team Member(s)
We are excited to welcome Matthias Hinterndorfer as a new Postdoc in the team and Katharina Kladnik as a new TA (see lab members section). Moreover, we are happy to host Miquel Duran-Frigola as a guest scientist for the next couple of months.
Presentation
at the DFCI Targeted Protein Degradation Webinar Series is now online. Check out the video below
New perspective in Cell Chem Bio
Natalie, Cris and Georg collected their thoughts to review how multi-omics approaches can inform the identification and characterization of molecular glues and PROTACs.
Welcome, Fabian
Fabian Offensperger joins the team as a postdoc. Check out details in the Team Members section
New Review out in Molecular Cell
Congratulations to Martin for his review on “Fast-acting chemical tools to delineate causality in transcriptional control".
Welcome, Gianluca
We are excited to have Gianluca Degliesposti join our team, working on computational approaches to large scale chemoproteomic studies. (see "Lab Members" for more information)
New lab members
We are super excited to welcome Gary Tin (Postdoc, chemistry), Eleonora Barone (TA molecular biology) and Andrea Rukavina (TA, proteomics) to the team. Learn more about them in the “ Lab members” section.
Good Bye, Cris!
With a heavy heart, we said good bye to Cristina Mayor-Ruiz, who joined the lab back in 2017 as the first postdoc in the group. While it is sad to see her leave, we couldn't be prouder and more excited about her new gig as she is setting up her own lab at the IRB in Barcelona. Cristina's passion for research and dedication to success will be missed, but shall serve as a blueprint for all other postdocs that joined the group more recently.
New paper in Nature Chemical Biology
Graphical Abstract of our latest paper on the identification of Cyclin K molecular glue degraders
Our team, led by postdoc Cristina Mayor-Ruiz describes a scalable strategy to discover molecular glues that induce the degradation of Cyclin K via chemical profiling in hyponeddylated cells. Please find a quick primer below.
As a lab, we are convinced about the incredible therapeutic potential of molecular glue degraders, particularly considering delivering on the promise of degrading “undruggable” proteins. However, their discovery has thus far been largely serendipitous. To change this, we set out to generate hyponeddylated cell models where many CRL E3 ligases are functionally impaired. We coupled this cell lines to differential anti-proliferative screens and identified a set of compounds that appeared dependent on uninterrupted neddylation. As a proof of concept, we characterized a novel DCAF15-dependent glue degrader, dCeMM1, which induces the selective degradation of the splicing factor RBM39 following previously described mechanism of action. To characterize the mechanism of the additional three scaffolds, we conducted quantitative proteomics experiments, showing a profound downregulation of Cyclin K, an interaction partner of the transcriptional kinases CDK12/13. To identify the involved E3 ligase, we conducted a set of functional genomics experiments including genome-wide CRISPR screens and hybrid-capture based sequencing approach of drug-tolerant clones. This led us to identify a CUL4B ligase complex that appeared to be lacking a dedicated substrate receptor.
Coupling drug-pulldown assays with biochemical reconstitution experiments led us to identify a unique and therapeutically unexplored mechanism where drug-binding prompts dimerization between E3 and CDK12, leading to degradation of CCNK. s.
Proxygen
Of note, this and other discovery approaches will be moved to Proxygen, a brand-new spin-out that was incorporated in May and recently also awarded the BI innovation prize. Proxygen is currently filling multiple positions here in Vienna. Check out details here:
New publication in Nature Genetics
Very often, we use multi-omics approaches to study and benchmark small molecules or small molecule degraders. This time, we turned things around and engineered degradable alleles of core components of the human Mediator complex, a central molecular machine involved in gene control. We couple acute Mediator degradation with global approaches to study the ensuing consequences on nascent transcription, chromatin structure and chromatin composition. Got interested? Read the paper that was masterfully championed by PhD student Martin Jäger with support from colleagues at the MPI in Göttingen and Berlin.
New lab members..continued…
Welcome Natalie Scholes who joins our group to study glue degraders and other non-obvious ligand-induced degradation mechanisms via high-content chemical screening and systems biology (“members” section to be updated soon)
New lab members!
Welcome Ruzanna and Marko, joining our lab as postdoctoral researchers to work on chemical proteomics, targeted protein degradation and molecular glues (see lab members for details)
Innovation funds call positively reviewed
We are excited that our application for the innovation funds of the Austrian Academy of Sciences was reviewed positively and we are looking forward to using targeted protein degradation to understand transcriptional condensates in cancer.
New TPD Review
on challenges in targeted protein degradation out in Current Opinions in Chemical Biology (see link below)
FFG-Bridge Grant funded
We are very grateful that the FFG has funded an acedemic/industry collaboration with our colleagues at allcyte, geared to find novel degraders at single-cell resolution by drug screening directly in patient samples. Stay tuned.
CeMM enters into research collaboration on targeted protein degradation with Pfizer Inc.
A highly interesting research collaboration, coordinated by Georg and involving the Winter lab alongside other Research labs at CeMM. Learn more in the official press release.
ERC Starting Grant awarded !!
We are very excited that the lab has been awarded a prestigious Starting Grant from the European Research Commission (ERC). The project “Glue2Degrade” will be funded with 1.3 € million over a period of five years.
Publication in Molecular Cell
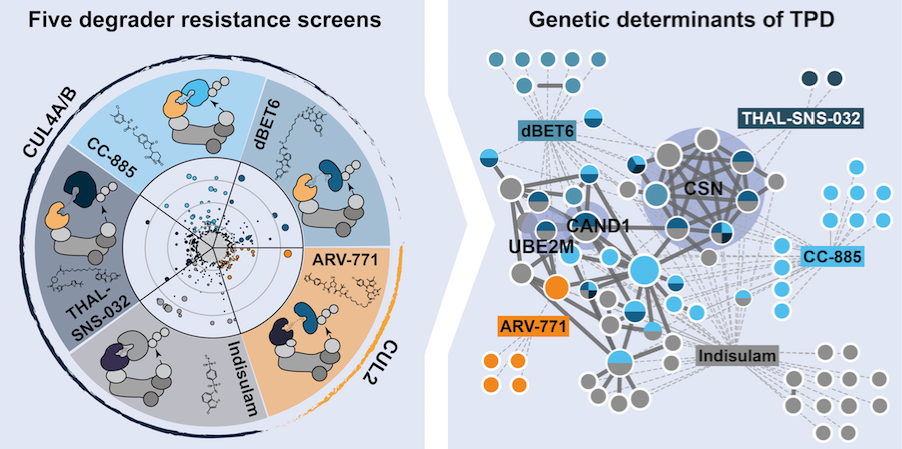
Cristina and Martin set out to understand all cellular effectors that are required for targeted protein degradation. Combining genome-scale CRISPR/Cas9 screens with quantitative proteomics, they uncovered global and specific effectors, and showed that perturbing the CAND1/CSN axis abrogates CRL plasticity and prompts ligase auto-degradation in a cell- and context-specific manner.
To find out more, read the paper ;)
News & Views in Nature Chem Biology
Matthias and Georg shared their thoughts on two excellent papers by the groups of Ben Cravatt and Dan Nomura. Both studies identified covalent ligands of E3 ubiquitin ligases (RNF114 and DCAF16) that are compatible with downstream PROTAC development. Particularly interesting, hijacking DCAF16 allows for a nuclear-specific target protein degradation.
Two New Grants Supporting our Research
We are very grateful for two recent Stand-Alone grants from the FWF that support our research at the interface of targeted protein degradation and gene control& oncogenic signal transduction.
Eppendorf Award 2019
Georg won the Eppendorf Award for Young European Investigators, acknowledging his contributions to the exciting field of targeted protein degradation. What a tremendous honor to follow so many of our scientific heros that won that award in previous years.
Protocol for engineering degradable alleles
Matthias outlined a detailed protocol for the generation of dTAG knock-ins. An easy to follow “to-do” list to get from the first primer to an endogenously tagged gene.
New Paper in Cell Chemical Biology
We teamed up with the lab of Nathanael Gray (DFCI) to develop and characterize a homolog-selective PROTAC for the cancer-target CDK6. Congratulations to our PhD student Matthias for his first first-author manuscript. Many more to follow!
Review on Targeted Protein Degradation
Cristina and Georg shared their views on the value of targeted protein degradation for target validation studies in cancer. This review is part of a special issue on targeted protein degradation with contributions from many friends and esteemed colleagues
Latest Tweets by Georg
-
Very exciting news from Proxygen, announcing a strategic collaboration with a deal size of up to $2.55 billion with… https://t.co/lYAwfrEasq
-
Very happy we could contribute to this exciting story out of the @laraialab. a very cool mechanism of action study… https://t.co/wHfOggU1NF
-
RT @alessiociulli: PLease RT! We have 4x opening at postdoctoral level to join our new @CeTPD_Dundee. Covering all range of discipline… https://t.co/O1JK4W5GMW
-
RT @clausen_lab: Join us at the "Ubiquitin & Friends" symposium in Vienna for exciting research in relaxed atmosphere. The hidden ge… https://t.co/XJPU4834hL
-
RT @LabThoma: Delighted to share our spotlight on a beautiful paper by @alessiociulli and @georg_e_winter. Alex Hanzl and colleag… https://t.co/NsMPwLnlTW
What defines our research?
Thematically, research in my lab is focused on the interface of cancer, chemical biology and gene control. We aim to innovate novel (pharmacologic) strategies that allow us to probe, understand and eventually target aberrant transcriptional circuits in cancer. Our science is thus unapologetically translational, but we nevertheless aim to ask scientific questions that are fundamental in nature and allow us to develop a better understanding of gene control and the ubiquitin-proteasome system. Our research strategy is inspired and driven by high-throughput and unbiased technologies such as quantitative proteomics, (nascent) transcriptomics and functional genomics. Integrative use of these technologies enables us to understand the mechanism of action of proteins, protein complexes or small molecules on a holistic level. In almost all cases, we are however not satisfied by merely charting the global picture, but rather use this multi-omics approaches to establish hypotheses that are followed up on a mechanistic level. Currently, the main focus of the group is to push the limits of targeted protein degradation, and -in the wider sense- proximity-inducing drugs, as a novel paradigm in therapeutic discovery.
multidisciplinary | technology enabled | mechanistically oriented | therapeutically inspired
We are a multi-disciplinary research lab interested in chemical approaches to control gene activity in cancer.
Targeted Protein Degradation
Leaving the paradigm of pharmacologic inhibition.
adapted from Nair et al., Cell 2003,PDB1NKP
Concepts in the design of small-molecule drugs continue to follow a predictable path. Scientists typically aim to identify high-affinity binders of hydrophobic pockets to inhibit the activity of a target protein. While successful for enzymes or receptors, this inhibitor-centric paradigm forces scientists to seek innovation where it is easiest to look for. Consequently, 80% of the proteome, including some of the most compelling disease-relevant targets such as MYC, RAS, or b-catenin, remain beyond the reach of therapeutic development.
Our approach towards blocking otherwise “undruggable” proteins is to hijack the ubiquitin-proteasome system via small-molecules in order to chemically “earmark” these proteins for degradation by the proteasome. Targeted protein degradation is based on small-molecules, generally called “degraders”, that induce the degradation of proteins by modulating the ubiquitination activity of E3 ligases. Overall, there are two types of small-molecule degraders, both of which are of interest to us: (i) non-chimeric compounds that act as molecular glues, or (ii) hetero-bifunctional molecules, often referred to as “proteolysis-targeting chimeras“ (PROTACs). Both function by inducing associations of a target protein of interest (POI) and an E3 ubiquitin ligase. Induced molecular proximity consequently prompts ubiquitin transfer to the POI and its ensuing proteasomal degradation.
Molecular Glues | MG
Examples of molecular glues include thalidomide, lenalidomide and the related family of “immunomodulatory drugs” (IMiDs), certain aryl sulfonamides, as well as several plant hormones, including auxin. All of these small-molecules bind substrate receptors (SR) of the cullin RING (CRL) family of E3 ligases. Binding induces or complements a neomorphic interaction surface that leads to the cooperative binding of a given neo-substrate, ubiquitin transfer, and its subsequent degradation by the proteasome. Intriguingly, many of these MGs induce the degradation of transcriptional regulators that would otherwise be considered as “undruggable”. Unfortunately, the discovery of MGs has been a process entirely driven by serendipity, and no rational discovery strategies exist.
The phytohormone auxin, a naturally occuring molecular glue. Tan X. et al, Nature 2007. PDB:2PN1
We are convinced that MGs could emerge as a key therapeutic strategy towards disrupting gene-regulatory circuits in cancer. We have identified scalable tools and phenotypic screens to identify novel MGs. In the lab, we are using functional genetic screens and quantitative proteomics approaches to further characterize novel MGs. Upon mechanistic characterization, the goal is to further optimize identified MGs by the means of medicinal chemistry to develop innovative, well-characterized chemical probes that might represent a vector for ensuing drug-development programs outside of academia.
Heterobifunctional Degraders | PROTACs
PROTACs are based on a modular design where a targeting warhead and an E3-ligase binder are connected via a flexible linker. PROTACs can thus simultaneously bind to an E3 ligase and a POI, enforce their molecular proximity, and induce POI ubiquitination and degradation. Due to this modular architecture, PROTACs can in principle be adapted to different POIs simply by exchanging the targeting warhead. This potential versatility rendered PROTACs particularly interesting for the pharmaceutical development, and led to a recent increase in the number of probes for preclinical target exploration.
We have recently reported the first in vivo compatible heterobifunctional degrader (Winter et al. Science 2015). Conjugation of a competitive BET-bromodomain antagonist to an IMiD-like phthalimide moiety afforded the heterobifunctional compound dBET1. dBET1 induced fast, potent and selective degradation of BET proteins BRD2, BRD3 and BRD4 in cell lines and in mouse xenograft experiments, and continuous degrader treatment outperformed competitive BET inhibition in disseminated AML xenografts. We furthermore showed that phthalimide conjugation is a generalizable approach by additionally reporting heterobifunctional degraders of the prolyl isomerase FKBP12. More recently, it has emerged that converting a small-molecule binder into a heterobifunctional degrader can offer means to engineering selectivity in otherwise promiscuous chemical scaffolds. In a recent manuscript, we teamed up with researchers at the Dana Farber Cancer Institute to describe novel degraders that allow the homolog-selective degradation of the kinase CDK6, a prolific target for the treatment of Acute Myeloid Leukemia (Brand et al., Cell Chem Bio 2018).
Gene Control in Cancer
Aberrant gene-control is a multifaceted key feature of human cancer. Dysregulated transcriptional circuits can arrest cancer cells in an undifferentiated, proliferative state by preventing definitive lineage commitment. In other instances, aberrant transcriptional networks might prompt the continuous overexpression of an oncogene, cause a dormant, highly drug-resistant cell state, or cause a primary tumor cells to colonize distant organs. Despite the paramount importance of transcription for arguably all key aspects of cancer, the spectrum of drugs capable to block or remodel aberrant gene control is very limited.
Our lab is interested in understanding the molecular basis of aberrant gene-regulation in cancer, particularly focusing on blood cancers, but also on malignancies driven by mutant Ras proteins. Towards that end, we combine acute protein ablation systems (“degradation Tag”/ “dTAG”), or direct small-molecule inhibitors and degraders (PROTACs or MGs; see above) with global and unbiased measurements of gene activity and genome structure. To capitalize on the high kinetic resolution of protein degradation systems, our focus is on nascent RNA-sequencing methods. We believe that a quantitative understanding of fundamental aspects of human gene-control is key to the innovation of novel therapeutic strategies. Recent work has elucidated the role of BET proteins as master regulators of pause release (Winter et al., Mol Cell 2017), and identified the YEATS domain containing protein ENL as a therapeutic target to inhibit AML-specific transcriptional circuits (Erb et al., Nature 2017).
Chemical Genetics and ChemOProteomics
As a chemical-biology lab, the identification of protein targets of (anti-cancer) small-molecules, as well as the elucidation of their mechanism-of-action is our great passion. In our efforts to find and characterise novel molecular glues that degrade non-drugged cancer targets , we employ two sub-disciplines of chemical biology:
Chemical Genetics
To understand cellular effectors required for drug-action, or to identify the biologically relevant target of small-molecule drugs, we apply a combination of the various functional-genomics strategies, including genome-wide, pooled CRIPSR/Cas9 screens, NGS-based profiling of drug-resistant sub-clones, and cellular barcoding studies to monitor the evolution of drug-resistant clones.
Chemical proteomics
For the vast majority of small-molecules of synthetic or natural origin, we don’t understand their associated spectrum of target proteins. This is of particular relevance for small molecules that are identified through chemical screens with a phenotypic, cellular readout. Moreover, most small-molecules are much more promiscuous than anticipated. This means that they don’t only bind one protein in a cellular context, but might modulate a range of closely related, but also completely dissimilar protein targets. Towards functionally and mechanistically annotating small-molecules with a non-understood anti-cancer phenotype, we are employing target-identification assays that are based on chemical proteomics. In chemical proteomics, small-molecules of interest are immobilised on beads and exposed lysates of cells of interest. Proteins bound to the drug-affinity resin are subsequently identified via unbiased, quantitative proteomics, and validated via orthogonal strategies.
THE TEAM - Current Lab Members
Our team consists of research scientists, graduate students and postdoctoral researchers with a mixed background in molecular- and cell biology, chemistry and computational biology. Our lab is located at the Center for Molecular Medicine of the Austrian Academy of Sciences (CeMM).
Marko Cigler
Initially trained as a chemist in Zagreb, Marko completed his PhD in the lab of Kathrin Lang at the Technical University Munich where he worked on genetic code expansion. He joined the lab as a postdoctoral fellow in March 2020 to work on developing molecular glue degraders.
Matthias Hinterndorfer
Matthias did his PhD in Johannes Zuber’s lab at the Institute of Molecular Pathology Vienna, where he established temporally resolved CRISPR screens that led to the discovery of the vertebrate nuclear proteasome import pathway. In his work in the Winter lab, he is now looking to combine his interests in genetic screening and proteasome biology to elucidate the genetic determinants underlying chemically induced targeted protein degradation.
David Hoi
Initially trained as chemist, David completed his PhD in Molecular Biology at the Institute of Molecular Pathology in Vienna, where he studied novel targeted protein degradation strategies in pathogenic bacteria. He joined the Winter Lab in November 2023 aiming to develop chemical solutions to rewire transcriptional regulation in cancer. In his free time David is passionate about outdoor sports & tennis as well as cooking.
Chrysanthi Kagiou
Chrysanthi received her master's degree in Molecular Mechanisms of Disease at Radboud University in The Netherlands where she worked at Dr. Hobo's lab at Radboudumc, focusing on hematological malignancies. She completed her master thesis at Dr. Weigelin's group at Werner Siemens Imaging Center, in Germany studying cytotoxic T cells in melanoma. In September 2020, Chrysanthi joined Loizou Lab at the Institute of Cancer Research in Vienna, using advanced genome editing technologies to study cancer-associated mutations on DNA damage response genes. Chrysanthi joined Winter lab as a PhD student in June 2022, focusing on identifying novel molecular glue degraders in different cell states.
Lisa Kainacher
Lisa started as a PhD student in the Winter lab in autumn 2023. She studied Molecular Biotechnology in her BSc at the FH Campus Wien. For her undergraduate research, she joined the Christodoulou lab at University College London to investigate the structural underpinnings of protein misfolding. She then studied Molecular Medicine in her MSc at the University of Vienna. For her master research, she joined the Murray lab at the Max Planck Institute of Biochemistry in Munich where she worked on the complex role of tryptophan metabolism in human cancers and its association with ferroptotic cell death. In her PhD, Lisa will focus on the rewiring of transcriptional networks using small molecule libraries and chemical screens to exploit cancer vulnerabilities.
Katharina Kladnik
Katharina joined the Winter lab in 2022 as Technical Assistant. She completed her Masters studies in molecular biotechnology at the FH Campus Wien, where her interests focused around cancer and biopharmaceutical research. Besides working on targeted protein degradation, Katharina enjoys to play table tennis, travel the world and read.
Amanda Ng
Amanda joined the lab in September 2019. She completed her undergraduate studies in Fumio Motegi’s lab at the Temasek Life Sciences Laboratory and the National University of Singapore, where she studied the suppressors of PAR-1 in C. elegans
Fabian Offensperger
Fabian completed his studies in Life Science at the University of Konstanz. There he joined the lab of Martin Scheffner to work on the characterization of the ubiquitin ligase UBE3A/E6AP by small molecules during his PhD. In March 2021, Fabian started in the Winter lab as a postdoctoral fellow to work on chemical proteomics and assay development for ligand binding. In his free time he enjoys outdoor activities like biking, climbing and all types of skiing.
Miquel Muñoz i Ordono
Initially trained as a biochemist in Barcelona, Miquel conducted his Master studies in Molecular biology at Utrecht University. After completing his undergraduate studies in Anna Obenauf’s lab at the Institute for Molecular Pathology in Vienna, where he studied the role of dendritic cells in immunotherapy-resistant melanoma, he joined the Winter lab in September 2020.
Caroline Schätz
Caroline studied Medical and Pharmaceutical Biotechnology at the IMC University of Applied Sciences in Krems. For her bachelor thesis she spent seven months at the Harvard Medical School in Boston working on tissue engineering using hypoimmunogenic stem cells. During this time, she also received the Marshall Plan Fellowship. For her master thesis she was working on the development of advanced organoid systems in the group of Christoph Bock in Vienna. In October 2022 Caroline joined the Winter lab as a PhD student, where her research efforts focus on dissecting drug feedback mechanisms using genetic screens as well as chemical screens in organoids.
Natalie Scholes
Initially trained as a chemist at the LMU Munich, she completed her Masters and PhD studies at Imperial College London combining synthetic and systems biology to engineer de novo developmental patterning. Natalie joined the group in May 2020 to study glue degraders and other non-obvious ligand-induced degradation mechanisms via high-content chemical screening and systems biology.
Dmitri Segal
Dmitri completed his PhD in molecular genetics at the University of Toronto under the supervision of Dr. Mikko Taipale. There, he utilized high-throughput interaction proteomics and high-content immunofluorescent imaging to decipher functional differences between 14-3-3 paralogs and uncover their chaperone-like functions. Dmitri joined the Winter lab in June 2023 as a postdoctoral fellow to identify and characterize novel proximity inducing compounds that rewire the function of specific transcriptional/epigenetic regulators in cancer. In his free time, Dmitri enjoys the great Canadian pastime of playing hockey.
Gary Tin
Gary is a medicinal chemist from the University of Toronto with an interest in chemical biology. His PhD work, under the supervision of Patrick Gunning, entailed designing novel covalent warheads towards targeting oncogenic proteins. He joined the lab in Dec 2020 as a postdoctoral fellow to pursue novel mechanisms of E3 ligase recruitment by molecular glue degraders. In his spare time, he enjoys travelling to experience different cultures, cooking and baking, hiking and playing the piano.
Georg Winter
Georg did his PhD at CeMM with Giulio Superti-Furga, working on elucidating the mechanism of action of anti-neoplastic drugs. He specialized on proteomics- as well as chemical genetics approaches to identify drug resistance mechanisms and synergistic drug combinations. He continued his training in chemical biology, working as a postdoctoral fellow with Dr. James Bradner the Dana Farber Cancer Institute/Harvard Medical School. Supported by an EMBO fellowship, he published the first paper reporting on in vivo target protein degradation (Winter et al., Science 2015). His postdoctoral work led to the foundation of C4 Therapeutics, now listed at the NASDAQ. He was recruited as a CeMM Principal Investigator in June 2016 where his research is focused on using the unique pharmacology of targeted protein degradation to understand and disrupt aberrant gene control circuits in cancer His interdisciplinary research lab is supported by several national and international grants and fellowships including an ERC Starting Grant. Moreover, Georg is a coordinator of a research partnership between CeMM and Pfizer. His work on molecular glue degraders led to the incorporation of Proxyen in May 2020. His work on PROTACs contributed to the incorporation of Solgate Therapeutics. Dr. Winter’s contribution to the field of targeted protein degradation was acknowledged via multiple prices and awards, including the Wilson S. Stone Memorial Award from MD Anderson (2021), the Eppendorf Award 2019 the Elisabeth Lutz Award of the Austrian Academy of Sciences.
Winter lab Alumi
HANA IMRICHOVA (2018-2023)
Hana was leading most computational efforts in her lab and was the master of all things -NGS.
MARK KUDADY (2022-2023)
Mark worked on approaches to understand hydrophobic tagging via engineering FACS-based stability reporters amenable to pooled CRISPR/Cas9 screens
ALEX HANZL (2018-2023)
Alex was the 3rd PhD student to join (and graduate) from the lab. He focused his efforts on mapping functional hotspots in E3 ligases that can be predictive for resistance acquisitions (Hanzl, Nat Chem Biol 2023). In addition, he developed a novel approach to identify molecular glue degraders in a ligase-centric fashion (Hanzl, JACS 2023). Alex will continue his scientific path in the Thomä lab where he will start as a postdoc in mid of 2023.
ANNA SCHREMPF (2022)
Anna joined our group just for a couple of month to finish her PhD studies. She quickly became a highly valued and integrated team member and we wish her all the best for her next career steps.
VINCENTH BRENNSTEINER (2021-2022)
Vincenth worked as a junior datascientist and was responsible for handling, processing and analyzing large-scale chemoproteomics datasets. He also gained additional and highly valuable experience in machine learning and artificial intelligence. Since 2023, he is fully employed by CeMM's proteomics facility.
ANDREA RUKAVINA (2020-2022)
Andrea worked as a TA at the interface of our research group and CeMM's proteomics facility, especially focusing on our industry collaboration. Andrea gained a lot of additional experience in (chemical) proteomics and mass spectrometry and has since then joined the proteomics facility.
SARAH DOBNER (2020-2022)
Sarah joined the team to work as at TA on an industry collaboration focused on large-scale chemoproteomics. She quickly became an indispensable team member and we will miss her dedication and attention to detail. We are however very excited that she is embarking on her PhD journey and wish her all the best!
ELEONORA BARONE (2020-2022)
Eleonora worked as a TA in the group, focusing on several TPD projects. In particular, she was working on assays to support the characterization of novel CRBN modulators. Moreover, she also critically supported efforts towards validating hypotheses we derived from deep mutational scanning data. Eleonora will continue her scientific journey by starting her PhD studies in the near future.
Elisa Hahn (2019-2022)
Elisa worked as a TA in the group, contributing tremendously important work in implementing and scaling our chemoproteomics capabilities. Elisa has moved on to continue her PhD studies, and will be dearly missed in the Winter lab. Not only for her excellent work, but also for her integrative, charming and upbeat character and spirits.
Martin Jäger (2016-2021)
Martin was a PhD student in the lab with a keen interest at the interface of chemical biology and gene control. He focused his efforts on understanding the acute role of the human Mediator complex in transcription, and performed genome-scale CRISPR/Cas9 screens to understand the genetic determinants of targeted protein degradation. In fall 2021, Martin started his postdoctoral studies in Ben Cravatt's group at the Scripps.
SOPHIE BAUER (2016-2021)
Sophie was the first person to join the group and hence instrumental in setting up the lab organisation and establishing a joyful working atmosphere. She made significant contributions to numerous publications, including the discoveries of selective CDK6 PROTACs and Cyclin K degraders. During her time in the lab, she became an expert in functional genomics, particularly CRISPR/Cas9 screens. We miss Sophie, but are excited for her new position as scientist at Proxygen.
Cristina Mayor-Ruiz (2017-2020)
Cristina conducted her postdoctoral studies in the Winter lab, aiming to determine the genetic requirements of targeted protein degradation to (i) understand potential resistance mechanisms, and (ii) deduce strategies for the discovery of novel molecular glue degraders. After 3 very successful years, Cris moved back to Spain to start her independent research lab at the IRB in Barcelona. Congrats!!
Ruzanna Mnatsakanyan (2020)
Ruzanna worked as a project scientist and implemented a novel workflow and pipeline for chemical proteomics.
Matthias brand (2016-2020)
Matthias performed his PhD studies in the Winter lab from 2016-2020. He worked on tuning the selectivity of PROTACs to develop homolog-selective degraders. Moreover, he was interested in charting resistance mechanisms to targeted protein degradation via functional genomic approaches.
Matthias has graduated in 2020 and co-founded Proxygen, a CeMM spin-out company that is dedicated to the scalable discovery of molecular glue degraders.
Diogo Borges Lima (2020)
In 2020, Diogo worked as a data scientist in the Winter lab, helping to develop chemoproteomic workflows
SOMETH CHORN (2016-2017)
Someth was a Master’s student working on off-target mechanisms of CRBN-based PROTACs. His studies resulted in novel insights into off-target mechanisms of protein degraders and a co-first author publication in ACS Chem Bio. Someth moved on to work at Boehringer Ingelheim in Vienna.
Publications
(lab members are highlighted in bold)
2023
Kozicka Z, Suchyta DJ, Focht V, Kempf G, Petzold G, Jentzsch M, Zou C, Di Genua C, Donovan KA, Coomar S, Cigler M, Mayor-Ruiz C, Schmid-Burgk JL, Häussinger D, Winter GE, Fischer ES, Słabicki M, Gillingham D, Ebert BL, Thomä NH. Design principles for cyclin K molecular glue degraders. Nature Chemical Biology 2023 Sep 7. doi: 10.1038/s41589-023-01409-z. Online ahead of print.
Enders L, Siklos M, Borggräfe J, Gaussmann S, Koren A, Malik M, Tomek T, Schuster M, Reiniš J, Hahn E, Rukavina A, Reicher A, Casteels T, Bock C, Winter GE, Hannich JT, Sattler M, Kubicek S. Pharmacological perturbation of the phase-separating protein SMNDC1. Nature Communications 2023 Aug 16;14(1):4504. doi: 10.1038/s41467-023-40124-0.
Dvorak V, Casiraghi A, Colas C, Koren A, Tomek T, Offensperger F, Rukavina A, Tin G, Hahn E, Dobner S, Frommelt F, Boeszoermenyi A, Bernada V, Hannich JT, Ecker GF, Winter GE, Kubicek S, Superti-Furga G. Paralog-dependent isogenic cell assay cascade generates highly selective SLC16A3 inhibitors. Cell Chemical Biology 2023 Aug 17;30(8):953-964.e9. doi: 10.1016/j.chembiol.2023.06.029. Epub 2023 Jul 28.
Nianzhe He, Laura Depta, Cecilia Rossetti, Marko Cigler, Marine Michon, Oliver Rafn Dan, Joseph Hoock, Julien Barbier, Daniel Gillet, Alison Forrester, Georg E. Winter, Luca Laraia. Selective inhibition of OSBP blocks retrograde trafficking by inducing partial Golgi degradation. biorxiv. April 2.
Marko Cigler, Hana Imrichova, Fabian Frommelt, Laura Depta, Andrea Rukavina, Chrysanthi Kagiou, J. Thomas Hannich, Cristina Mayor-Ruiz, Giulio Superti-Furga, Sonja Sievers, Luca Laraia, Herbert Waldmann# and Georg E. Winter#. Orpinolide disrupts a leukemic dependency on cholesterol transport by inhibiting the oxysterol-binding protein OSBP. biorxiv. March 15
Gang Xue, Jianing Xie, Matthias Hinterndorfer, Marko Cigler, Hana Imrichova, Soheila Rezaei Adariani, Lara Dötsch, Georg E. Winter# and Herbert Waldmann#.Discovery of a Drug-like, Natural Product-Inspired DCAF11 Ligand Chemotype. chemrxiv. March 10.
Oliver Hsia, Matthias Hinterndorfer, Angus D. Cowan, Kentaro Iso, Tasuku Ishida , Ramasubramanian Sundaramoorthy , Mark A. Nakasone , Andrea Rukavina , Koraljka Husnjak , Martin Wegner, Alejandro Correa-Sáez, Conner Craigon, Chiara Maniaci, Andrea Testa, Manuel Kaulich, Ivan Dikic , Georg E. Winter# and Alessio Ciulli#. An intramolecular bivalent degrader glues an intrinsic BRD4-DCAF16 interaction. biorxiv. 2023 Feb 14.
Duran-Frigola M, Cigler M, Winter GE. Advancing Targeted Protein Degradation via Multiomics Profiling and Artificial Intelligence. J Am Chem Soc. Epub 2023 Jan 27.
Hanzl A, Barone E, Bauer S, Yue H, Nowak RP, Hahn E, Pankevich EV, Koren A, Kubicek S, Fischer ES, Winter GE. E3-Specific Degrader Discovery by Dynamic Tracing of Substrate Receptor Abundance. J Am Chem Soc. 2023 Jan 5.
Tin G, Winter GE. CIDE-stepping E3s. Nat Chem Biol. 2023 Jan;19(1):3-4.
2022
Schrempf A, Bernardo S, Arasa Verge EA, Ramirez Otero MA, Wilson J, Kirchhofer D, Timelthaler G, Ambros AM, Kaya A, Wieder M, Ecker GF, Winter GE, Costanzo V, Loizou JI. POLθ processes ssDNA gaps and promotes replication fork progression in BRCA1-deficient cells. Cell Rep. 2022 Nov 29;41(9):111716.
Hanzl A, Casement R, Imrichova H, Hughes SJ, Barone E, Testa A, Bauer S, Wright J, Brand M, Ciulli A, Winter GE. Functional E3 ligase hotspots and resistance mechanisms to small-molecule degraders. Nat Chem Biol. 2022 Nov 3.
Peter B, Eisenwort G, Sadovnik I, Bauer K, Willmann M, Rülicke T, Berger D, Stefanzl G, Greiner G, Hoermann G, Keller A, Wolf D, Čulen M, Winter GE, Hoffmann T, Schiefer AI, Sperr WR, Zuber J, Mayer J, Valent P. BRD4 degradation blocks expression of MYC and multiple forms of stem cell resistance in Ph+ chronic myeloid leukemia. Am J Hematol. 2022 Sep;97(9):1215-1225.
2021
Müller S., Ackloo S, ..Winter GE, Workman P, Arrowsmith CH. Target 2035 - update on the quest for a probe for every protein. RSC Med Chem. 2021 Dec 3;13(1):13-21.
Zumer K, Maier KC, Farnung L, Jaeger MG, Rus P, Winter G, Cramer P. Two distinct mechanisms of RNA polymerase II elongation stimulation in vivo. Molecular Cell. 2021 Aug 5;81(15):3096-3109.e8.
Scholes NS, Mayor-Ruiz C, Winter GE. Identification and selectivity profiling of small-molecule degraders via multi-omics approaches. Cell Chemical Biology, 2021 Mar 29:S2451-9456(21)00146-X. doi: 10.1016/j.chembiol.2021.03.007. Online ahead of print.
Kiely-Collins H, Winter GE, Bernardes GJL. The role of reversible and irreversible covalent chemistry in targeted protein degradation. Cell Chemical Biology, 2021 Mar 25: S2451-9456(21)00144-6. doi: 10.1016/j.chembiol.2021.03.005. Online ahead of print.
Jaeger MG, Winter GE. Fast-acting chemical tools to delineate causality in transcriptional control. Molecular Cell, 2021 Mar 4:S1097-2765(21)00125-8. doi: 10.1016/j.molcel.2021.02.015. Online ahead of print
Schick S, Grosche S, Kohl KE, Drpic D, Jaeger MG, Marella NC, Imrichova H, Lin JG, Hofstätter G, Schuster M, Rendeiro AF, Koren A, Petronczki M, Bock C, Müller AC, Winter GE, Kubicek S. Acute BAF perturbation causes immediate changes in chromatin accessibility. Nature Genetics, 2021 Feb 8. doi: 10.1038/s41588-021-00777-3.
2020
Mayor-Ruiz C, Bauer S, Brand M, Kozicka Z, Siklos M, Imrichova H, Kaltheuner IH, Hahn E, Seiler K, Koren A, Petzold G, Fellner M, Bock C, Müller AC, Zuber J, Geyer M, Thomä NH, Kubicek S, Winter GE. Rational discovery of molecular glue degraders via scalable chemical profiling. Nature Chemical Biology, 2020 Aug 3. doi: 10.1038/s41589-020-0594-x
Berger M, Knittl-Frank C, Bauer S, Winter GE, Maulide N. Application of Relay C-H Oxidation Logic to Polyhydroxylated Oleanane Triterpenoids. Chem 2020, https://doi.org/10.1016/j.chempr.2020.04.007
Jaeger MG, Schwalb B, Mackowiak SD, Velychko T, Hanzl A, Imrichova H, Brand M, Agerer B, Chorn S, Nabet B, Ferguson FM, Müller AC, Bergthaler A, Gray NS, Bradner JE, Bock C, Hnisz D, Cramer P, Winter GE. Selective Mediator dependence of cell-type-specifying transcription. Nature Genetics. 2020 Jun 1. doi: 10.1038/s41588-020-0635-0. Online ahead of print.
Bensimon A, Pizzagalli MD, Kartnig F, Dvorak V, Essletzbichler P, Winter GE, Superti-Furga G. Targeted degradation of SLC Transporters reveals amenability of multi-pass transmembrane proteins to ligand-induced proteolysis. Cell Chemical Biology. 2020 Apr 18. pii: S2451-9456(20)30117-3. doi: 10.1016/j.chembiol.2020.04.003.
Hanzl A, Winter GE. Targeted protein degradation: current and future challenges. Current Opinion in Chemical Biology. 2020 Jan 2;56:35-41. doi: 10.1016/j.cbpa.2019.11.012.
2019
Ferreira da Silva J, Salic S, Wiedner M, Datlinger P, Essletzbichler P, Hanzl A, Superti-Furga G, Bock C, Winter G, Loizou JI. Genome-sale CRISPR screens are efficient in non-homologous end-joining deficient cells. Sci Rep. 2019 Oct 31;9(1):15751
Mayor-Ruiz C, Jaeger MG, Bauer S, Brand M, Sin C, Hanzl A, Mueller AC, Menche J, Winter GE. Plasticity of the Cullin-RING Ligase Repertoire Shapes Sensitivity to Ligand-Induced Protein Degradation. Molecular Cell. 2019 Aug 22;75(4):849-858.e8. doi: 10.1016/j.molcel.2019.07.013.
Brand M, Winter GE. Stick it to E3s. Nature Chemical Biology. 2019. Jul;15(7):655-656. doi: 10.1038/s41589-019-0312-8.
Mayor-Ruiz C, Winter GE. Identification and characterization of cancer vulnerabilities via targeted protein degradation. Drug Discovery Today. 2019 Apr;31:81-90. doi: 10.1016/j.ddtec.2018.12.003
Sdelci S, Rendeiro AF, Rathert P, You W, Lin JG, Ringler A, Hofstätter G, Moll HP, Gürtl B, Farlik M, Schick S, Klepsch F, Oldach M, Buphamalai P, Schischlik F, Májek P, Parapatics K, Schmidl C, Schuster M, Penz T, Buckley DL, Hudecz O, Imre R, Wang SY, Maric HM, Kralovics R, Bennett KL, Müller AC, Mechtler K, Menche J, Bradner JE, Winter GE, Klavins K, Casanova E, Bock C, Zuber J, Kubicek S. MTHFD1 interaction with BRD4 links folate metabolism to transcriptional regulation. Nature Genetics. 2019 Jun;51(6):990-998. doi: 10.1038/s41588-019-0413-z.
Brand M, Winter GE. Locus-Specific Knock-In of a Degradable Tag for Target Validation Studies. Methods Mol Biol. 2019;1953:105-119. doi: 10.1007/978-1-4939-9145-7_7.
2018
Brand M, Jiang B, Bauer S, Donovan KA, Liang Y, Wang ES, Nowak RP, Yuan JC, Zhang T, Kwiatowski N, Mueller AC, Fischer ES, Gray NS, Winter GE. Homolog-Selective Degradation as a Strategy to Probe the Function of CDK6 in AML, Cell Chem Biol. 2018 Nov (Epub ahead of print). PMID: 30595531
Bigenzahn JW, Collu GM, Kartnig F, Pieraks M, Vladimer GI, Heinz LX, Sedlyarov V, Schischlik F, Fauster A, Rebsamen M, Parapatics K, Blomen VA, Müller AC, Winter GE, Kralovics R, Brummelkamp TR, Mlodzik M, Superti-Furga G. LZTR1 is a regulator of RAS ubiquitination and signaling. Science. 2018 Dec; 362(6419):1171-1177. PMID: 30442766
Ishoey M, Chorn S, Singh N, Jaeger MG, Brand M, Paulk J, Bauer S, Erb MA, Parapatics K, Müller AC, Bennett KL, Ecker GF, Bradner JE, Winter GE. Translation Termination Factor GSPT1 Is a Phenotypically Relevant Off-Target of Heterobifunctional Phthalimide Degraders. ACS Chem Biol. 2018 Mar 16;13(3):553-560. PMID:29356495
Moser SC, Voerman JSA, Buckley DL, Winter GE, Schliehe C. Acute Pharmacologic Degradation of a Stable Antigen Enhances Its Direct Presentation on MHC Class I Molecules. Front Immunol. 2018 Jan 8;8:1920. PMID: 29358938
Xu L, Chen Y, Mayakonda A, Koh L, Chong YK, Buckley DL, Sandanaraj E, Lim SW, Lin RY, Ke XY, Huang ML, Chen J, Sun W, Wang LZ, Goh BC, Dinh HQ, Kappei D, Winter GE, Ding LW, Ang BT, Berman BP, Bradner JE, Tang C, Koeffler HP. Targetable BET proteins- and E2F1-dependent transcriptional program maintains the malignancy of glioblastoma. Proc Natl Acad Sci USA. 2018. E5086-E5095, vol 115, no. 22, PMID: 29764999
Nabet B, Roberts JM, Buckley DL, Paulk J, Dastjerdi S, Yang A, Leggett AL, Erb MA, Lawlor MA, Souza A, Scott TG, Vittori S, Perry JA, Qi J, Winter GE, Wong KK, Gray NS, Bradner JE. The dTAG system for immediate and target-specific protein degradation. Nat Chem Biol. 2018 May;14(5):431-441. PMID: 29581585
Gechijian LN, Buckley DL, Lawlor MA, Reyes JM, Paulk J, Ott CJ, Winter GE, Erb MA, Scott TG, Xu M, Seo HS, Dhe-Paganon S, Kwiatkowski NP, Perry JA, Qi J, Gray NS, Bradner JE. Functional TRIM24 degrader via conjugation of ineffectual bromodomain and VHL ligands. Nat Chem Biol. 2018 Apr;14(4):405-412. PMID: 29507391
Zeid R, Lawlor MA, Poon E, Reyes JM, Fulciniti M, Lopez MA, Scott TG, Nabet B, Erb MA, Winter GE, Jacobson Z, Polaski DR, Karlin KL, Hirsch RA, Munshi NP, Westbrook TF, Chesler L, Lin CY, Bradner JE. Enhancer invasion shapes MYCN-dependent transcriptional amplification in neuroblastoma. Nat Genet. 2018 Apr;50(4):515-523. PMID:29379199
Olson CM, Jiang B, Erb MA, Liang Y, Doctor ZM, Zhang Z, Zhang T, Kwiatkowski N, Boukhali M, Green JL, Haas W, Nomanbhoy T, Fischer ES, Young RA, Bradner JE, Winter GE, Gray NS. Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nat Chem Biol. 2018 Feb;14(2):163-170. PMID: 29251720
2017
Huang HT, Seo HS, Zhang T, Wang Y, Jiang B, Li Q, Buckley DL, Nabet B, Roberts JM, Paulk J, Dastjerdi S, Winter GE, McLauchlan H, Moran J, Bradner JE, Eck MJ, Dhe-Paganon S, Zhao JJ, Gray NS. MELK is not necessary for the proliferation of basal-like breast cancer cells. Elife. 2017 Sep 19;6. pii: e26693. PMID: 28926338
Winter GE, Mayer A, Buckley DL, Erb MA, Roderick JE, Vittori S, Reyes JM, di Iulio J, Souza A, Ott CJ, Roberts JM, Zeid R, Scott TG, Paulk J, Lachance K, Olson CM, Dastjerdi S, Bauer S, Lin CY, Gray NS, Kelliher MA, Churchman LS, Bradner JE. BET Bromodomain Proteins Function as Master Transcription Elongation Factors Independent of CDK9 Recruitment. Mol Cell. 2017 Jul 6 67, 5–18. PMID: 28673542
Erb MA, Scott TG, Li BE, Xie H, Paulk J, Seo HS, Souza A, Roberts JM, Dastjerdi S, Buckley DL, Sanjana NE, Shalem O, Nabet B, Zeid R, Offei-Addo NK, Dhe-Paganon S, Zhang F, Orkin SH, Winter GE, Bradner JE. Transcription control by the ENL YEATS domain in acute leukaemia. Nature. 2017 Mar 9;543(7644):270-274. PMID: 28241139
Radic-Sarikas B, Halasz M, Huber KVM, Winter GE, Tsafou KP, Papamarkou T,Brunak S, Kolch W, Superti-Furga G. Lapatinib potentiates cytotoxicity of YM155 in neuroblastoma via inhibition of the ABCB1 efflux transporter. Sci Rep. 2017 Jun 8;7(1):3091. PMID: 28596528
Winter GE. Protein degradation: DCAFinating splicing. Nat Chem Biol. 2017 Jun;13(6):575-576
2016
Peter B, Winter GE, Blatt K, Bennett KL, Stefanzl G, Rix U, Eisenwort G, Hadzijusufovic E, Gridling M, Dutreix C, Hoermann G, Schwaab J, Radia D, Roesel J, Manley PW, Reiter A, Superti-Furga G, Valent P. Target interaction profiling of midostaurin and its metabolites in neoplastic mast cells predicts distinct effects on activation and growth. Leukemia. 2016 Feb;30(2):464-72. PMID: 26349526
2015
Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe-Paganon S, Bradner JE. DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 2015. Science 348, 1376-81. PMID: 25999370
Dixon SJ, Winter GE, Musavi LS, Lee ED, Snijder B, Rebsamen M, Superti-Furga G, Stockwell BR. Human Haploid Cell Genetics Reveals Roles for Lipid MetabolismGenes in Nonapoptotic Cell Death. ACS Chem Biol. 2015 Jul 17;10(7):1604-9. PMID: 25965523
2014-2010
Winter GE, Radic B, Mayor-Ruiz C, Blomen VA, Trefzer C, Kandasamy RK, Huber KVM, Gridling M, Chen D, Klampfl T, Kralovics R, Kubicek S, Fernandez-Capetillo O, Brummelkamp TR, Superti-Furga G. The solute carrier SLC35F2 enables YM155-mediated DNA damage toxicity. Nat Chem Biol. 2014 Sep;10(9):768-773. PMID: 25064833
Bulut-Karslioglu A, De La Rosa-Velázquez IA, Ramirez F, Barenboim M, Onishi-Seebacher M, Arand J, Galán C, Winter GE, Engist B, Gerle B, O'Sullivan RJ, Martens JH, Walter J, Manke T, Lachner M, Jenuwein T. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol Cell. 2014 Jul 17;55(2):277-90. PMID: 24981170
Li J, Bennett K, Stukalov A, Fang B, Zhang G, Yoshida T, Okamoto I, Kim JY, Song L, Bai Y, Qian X, Rawal B, Schell M, Grebien F, Winter GE, Rix U, Eschrich S, Colinge J, Koomen J, Superti-Furga G, Haura EB. Perturbation of the mutated EGFR interactome identifies vulnerabilities and resistance mechanisms. Mol Syst Biol. 2013 Nov 5;9:705. PMID: 24189400
Borgdorff V, Rix U, Winter GE, Gridling M, Müller AC, Breitwieser FP, Wagner C, Colinge J, Bennett KL, Superti-Furga G, Wagner SN. A chemical biology approach identifies AMPK as a modulator of melanoma oncogene MITF. Oncogene. 2014 May8;33(19):2531-9. PMID:23728343
Winter GE, Rix U, Carlson SM, Gleixner KV, Grebien F, Gridling M, Müller AC, Breitwieser FP, Bilban M, Colinge J, Valent P, Bennett KL, White FM, Superti-Furga G. Systems-pharmacology dissection of a drug synergy in imatinib-resistant CML. Nat Chem Biol. 2012 Nov;8(11):905-912. PubMed PMID: 23023260
Winter GE, Rix U, Lissat A, Stukalov A, Müllner MK, Bennett KL, Colinge J, Nijman SM, Kubicek S, Kovar H, Kontny U, Superti-Furga G. An integrated chemical biology approach identifies specific vulnerability of Ewing's sarcoma to combined inhibition of Aurora kinases A and B. Mol Cancer Ther. 2011 Oct;10(10):184656. PMID: 21768330
Ubaida Mohien C, Hartler J, Breitwieser F, Rix U, Remsing Rix L, Winter GE, Thallinger GG, Bennett KL, Superti-Furga G, Trajanoski Z, Colinge J. MASPECTRAS2: An integration and analysis platform for proteomic data. Proteomics. 2010 Jul;10(14):2719-22. PMID: 20455215
Where to find us.
CeMM -Research Center for Molecular Medicine of the Austrian Academy of Science
Lazarettgasse 14, AKH BT 25.3
1090 Vienna, Austria
How to contact us.
Interested in joining the lab?
Do you want to collaborate with us?
Do you need help with/access to tools we have generated?
Contact us: gwinter(at)cemm.at